GSoC - 1
This is the first blog that documents the coding period of my GSoC21 journey. I learnt a few interesting things in these two weeks, as I expected I would. So, let’s dive in and see if you knew few of these stuff I learnt.
Starting off !!!
I started off by getting a brief idea of the scope of the changes that could be done to the dataframe. This was the task I had decided on for the first week. Whenever we are involved in a project that runs for a period of anywhere between 2-4 months it is important to have a timeline or a roadmap of sorts to be able to look back to. This doesn’t really have to be something rigid. We can chose to deviate from it and infact deviations are bound to happen due to multiple reasons. It can happen because of an unexpected bug in between, or because you came across some alternative that you did not consider at the start or simply because it is one of those projects that gives better insights as you dwell into it.
Every good GSoC proposal consists of a tentative timeline that depicts the work we plan on doing as the weeks progress. Here is the timeline I had submitted in my proposal.
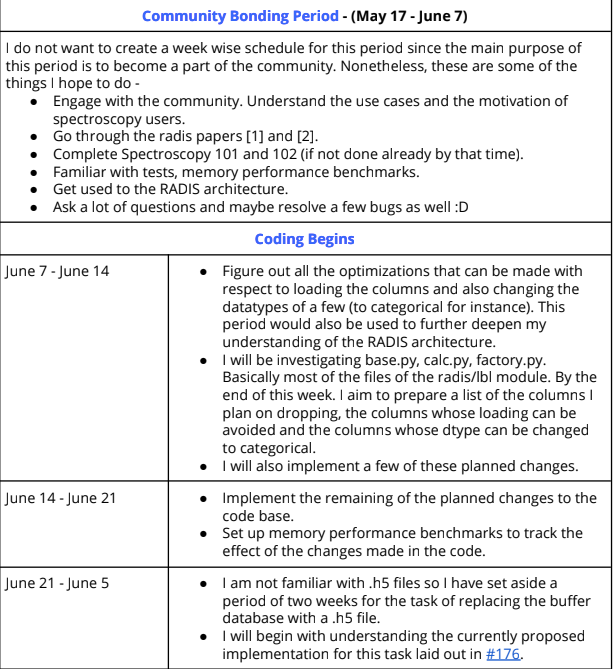
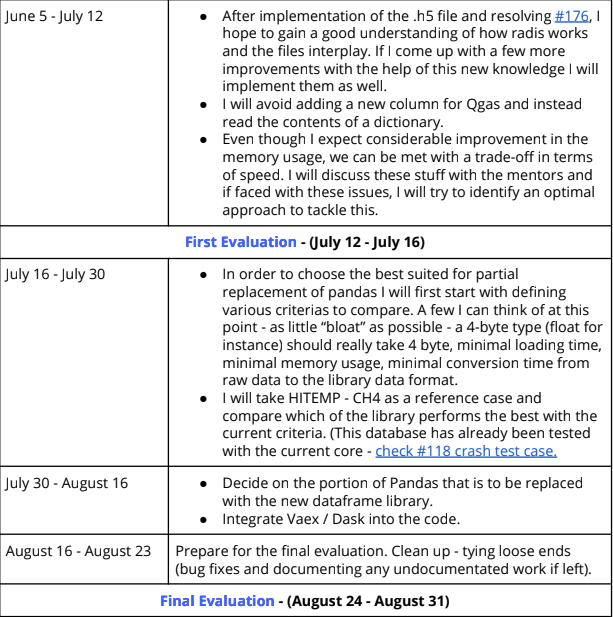
So as per this I was supposed to finish off the refactors to the dataframe and also finish setting up the benchamarks. But I was unable to complete these. I had underestimated the work it would take to complete them. Nonetheless, I also did have some time to look up at the things I am supposed to do in the second half of the coding period.
Memory and Time performance benchmarks - Tic-Tok
Before making any changes to the codebase Erwan suggested me to have the benchmarks setup. So what do I mean by this? To make sure that the changes I am making to the code are indeed reducing the memory consumption of the computations we use a few tools that help us track the memory consumption for various calculations as a function of git commits. There are multiple tools that help us do this. Radis already used a tool developed by airspeed velocity to track the memory computions. I ran into a lot of troubles in setting these up and a lost a lot of valuable time in the process ultimately Erwan fixed it and I was able to run the benchmarks on my machine.
The benchmarks still seem to take a lot of time to run though and for them to be feasible to be used a tool through which I can check the performance regularly there are a few things I need to learn. I hope to pick these up in the next few days.

We are also trying to look at a few other alternatives that can be used instead of asv. I will update the you guys regarding this in the next blog post.
Oh Pandas here I deal with you !
Let’s ditch a few columns
We can reduce the memory usage of pandas by using one really simple trick - avoid giving loading the columns that are not required for computation. Below I demostrate how just dropping a few columns can provide significant improvement in the memory consumption. I am using HITEMP-CH4 database for demonstration.
|
|
The peak memory usage before dropping the columns was 30.5 MB and once I remove a few columns the peak memory usage becomes 25.4 MB. I have already implemented the dropping of id column and handled the case of single isotope as well by dropping the column and istead just storing the information of the isotope as a meta attribute. We have also finalised on the discarding of the other columns by considering the physics of these quantities. Let’s check out a few of them. Since I haven’t already implemented the optimisations that follow I will save the implementation details for the next blog.
Einstein’s Coeffecients and Linestrengths
There are four parameters of interest to describe the intensity of a line : Linestrength $(int)$, Einstein emission coefficient $(A)$ and Einstein absorption coefificent $(B_{lu})$, Einstein induced emission coefficient $(B_{ul})$. All of them are somehow linked to the Squared Transition Dipole Moment $(R)$. 1
$$ B_{lu}=10^{-36}\cdot\frac{8{\pi}^3}{3h^2} R_s^2 \cdot 10^{-7} $$
$$ B_{ul}=10^{-36}\cdot\frac{8{\pi}^3}{3h^2} \frac{gl}{gu} R_s^2 \cdot 10^{-7} $$
$$ A_{ul}=10^{-36}\cdot\frac{\frac{64{\pi}^4}{3h} {\nu}^3 gl}{gu} R_s^2 $$
So now the idea would be to drop the $int$ column and use $A_{ul}$ to calculate the value of $int$ from it. The reason to drop $int$ and not $A_{ul}$ some databases like ExoMol databases only provide the value of $A_{ul}$.
Concat better
For anyone who wants concate multiple datafiles pandas tends to become useless as the memory scales up. I started out experimenting concat operations inorder to cluster the isotopes of each type, run computations on them and later concat them. But I later learnt that since this data is already in the form of a single dataframe, indexing is a better parameter to track the memory consumption. Nonetheless there are a few other places in Radis where we process multiple files and concat them, hence this experiment would help us decide how we can chose to replace the current approach with a better one. I tried out three methods. I was using some random dummy datafiles of around 780 MBs.
- Normal
pandas.concat - Concat with a doubly ended queue
- Concat with parquet
Here are the results of each of these methods -
pandas.concat
CPU Time - 0:02:43.797588
Peak Memory Usage - 4.1050 GB
pandas.concat with a doubly queue
CPU Time - 0:02:34.484612
Peak Memory Usage - 3.7725 GB
Concat with parquet
CPU Time - 0:01:37.984875
Peak Memory Usage - 1.6829 GB
Looking at the results, parquet seems like a really good option to me. But we will run for a few more examples and later check which one suits the best.
The next two weeks
The project is making progress in all fronts. I feel I need to reorganize my thoughts a bit. My main work for now would be to complete the task list of this pr. And then look at other stuff.