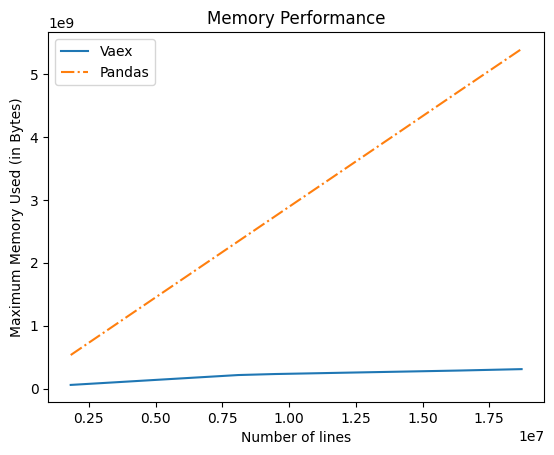
Comparing memory performance of Vaex and Pandas
After completing all the changes to compute Spectrum using Vaex , I compared the memory used during the execution of the program . I used tracemalloc to compute memory uses to compute Spectrum .
Computing the spectrum
Following code is used , and memory maximum memory used during the execution of this code is recorded for Vaex and Pandas
from radis import calc_spectrum
import tracemalloc
tracemalloc.start()
s, factory_s = calc_spectrum(1800, 2500, # cm-1
molecule='H2O',
isotope='1,2,3',
pressure=1.01325, # bar
Tgas=700, # K
mole_fraction=0.1,
path_length=1, # cm
databank='hitemp', # or 'hitemp', 'geisa', 'exomol'
wstep='auto',
use_cached=False,
engine='pandas',
return_factory=True,
)
s.apply_slit(0.5, 'nm') # simulate an experimental slit
s.plot('radiance')
print(tracemalloc.get_traced_memory())
tracemalloc.stop()
Results
It can be seen from the graph below that Vaex takes very less memory space in comparison to Pandas.