GSOC Week 1 : A global overview of the RADIS API.
GSOC Week 1 : A global overview of the RADIS API.
Since the point of my project is to implement new databases in the RADIS API and to reform it in other to make it common and stand alone, it seems important to understand how this API works.
Thus, I have started my 12 weeks projects by reading carefully the API documentation available at https://radis.readthedocs.io/en/latest/source/radis.io.dbmanager.html. I will detail in this article what I consider the key points to understand how Radis works globally and aim to answer the following question : Whats are the steps to proceed in order to include a new database as Kurucz to RADIS ?
This page is a description of the different modules available in the RADIS library except the api one which will be discussed later.
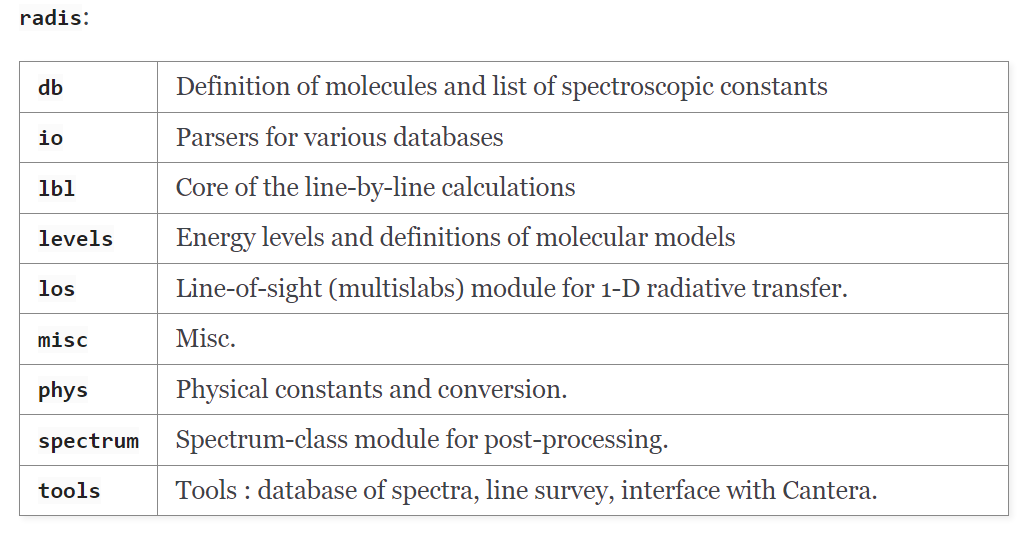
In particular, the radis.io package provided functionality for file management, downloading and analysis of different spectroscopic databases.
To understand in detail how the RADIS API works and integrate a new database like Kurucz, here are the most important radis.io package sub-modules to explore :
dbmanager : This submodule used to contain the DatabaseManager class, which is the heart of database management in RADIS before it was moved to the api module. Understanding how this class is used to save, download, manipulate and interact with existing databases is essential. I will also need to understand how to add a new database by implementing the appropriate methods.
geisa, hitran or exomol : I have to study how GEISA, HITRAN or Exomol files are downloaded, saved and processed. Understanding how this databases are integrated will give me an idea of the structure and workflow needed to integrate a new database.
The DatabaseManager class
The DatabaseManager class aims to handle and manage files from various databases. It provides a generic framework for managing and caching files from different sources or databases.
class DatabaseManager(object):
add_column(df, key, value): This method allows to create a column with a key and a value specified in a DataFrame or a dictionary.
check_deprecated_files(local_files, auto_remove=True): This method checks file metadata and removes deprecated ones. If auto_remove is set to True, deprecated files will be automatically removed. Otherwise, an error will be thrown.
clean_download_files(): This method cleans downloaded and unzipped files.
download_and_parse(urlnames, local_files): This method downloads and parses files from the specified URLs and saves them locally.
fetch_filenames(): This method fetches the names of all files from the database, even if they haven’t been downloaded yet.
fetch_urlnames(): This method must be overridden in the DatabaseManager subclass. It should return a list of URLs corresponding to database files.
get_columns(local_file): This method retrieves all the columns available in a database file using the get_columns function of the DataFileManager class.
load(local_files, columns=None, lower_bound=[], upper_bound=[], within=[], output=’pandas’): This method loads data from database files. You can specify which columns to load, lower and upper bounds for values in certain columns, and constraints for certain columns. You can also specify the output format, such as ‘pandas’, ‘vaex’ or ‘jax’.
plot(local_files, isotope, wavenum_min, wavenum_max): This method is a convenience function for plotting the linestrengths of the database.
register(dict_entries): This method registers dictionary entries for a specified database.
remove_local_files(local_files): This method removes the specified local files.
rename_columns(df, rename_dict): This method renames the columns of a DataFrame using a dictionary of correspondence between the old column names and the new names.
register_database(databank_name, dict_entries, verbose): This method adds registered databases to the RADIS configuration file.
The radis.io.geisa module review : to get some ideas ?
The GEISADatabaseManager class is a subclass of DatabaseManager which is specifically designed to manage GEISA databases. It adds specific functionalities to the management of GEISA files and their integration into the RADIS API. It gives us an insight of what the implemenation of other databases as Kurucz should look like. However, every database has its own specificities that is why the methods may differ on some aspects though they are built on a same basis.
fetch_urlnames(): This method must be implemented to return a list of URLs corresponding to GEISA files in your database. These URLs will be used to download files from the GEISA website.
parse_to_local_file(): This method is used to unpack GEISA files and save them locally. It also adds metadata to files.
register(): This method registers the GEISA database in the RADIS configuration file. It ensures that the RADIS API recognizes and can access the database.
fetch_geisa(): This GEISA specific method fetches GEISA files from the GEISA website, unpacks them and creates an HDF5 file containing all row data. It returns a Pandas DataFrame containing all rows.
columns_GEISA: It is a dictionary that defines the parsing order of columns in the GEISA2020 format. It specifies column names, data types, descriptions, and units.
engine: This is an optional parameter that allows to specify the memory mapping library to use for the GEISA database. By default, it uses the ‘default’ value specified in the RADIS configuration file.
verbose, chunksize, parallel: These are optional parameters to control the verbosity level of informational messages, the size of chunks for loading data and the use of parallel loading.
The fetch_geisa() method is an example of using the GEISADatabaseManager class to retrieve GEISA data for a specific molecule. This method downloads GEISA files from the GEISA website, unpacks them and saves them locally. It returns a Pandas DataFrame containing all rows from the GEISA database for the specified molecule.
Before exploring the Kurucz database structure more in details, let is discuss the other modules of the RADIS API.
The db module :
Definition of molecules: The db module allows to define the molecules used in the spectroscopic calculations. You can specify isotopes, molecular weights, chemical symbols, and other relevant molecular properties.
Spectroscopic constants: The db module stores the spectroscopic constants associated with each molecule, such as electronic transitions, energy levels, frequencies, absorption intensities, cross sections, etc.
Data Access: The db module provides methods and data structures to easily access information about molecules and spectroscopic constants.
Database extensions: The db module allows extending the functionalities of the database by adding new molecules, isotopes or spectroscopic constants. Here it is possible to add new data or integrate external databases to enrich the capabilities of RADIS.
Interoperability: The db module facilitates interoperability with other libraries and spectroscopic data formats. It allows to import and export spectroscopic data in different formats, such as HITRAN, HITEMP, GEISA, Kurucz, etc.
What about Exojax ?
Another open-source code, called EXOJAX, exchanged portions of code with RADIS until some parts of both codes became very similar. Currently, a database API is written in the RADIS code.
Understanding how Radis and Exojax somehow partially merged will be useful to understand how to implement a new database.
May 2022 : an example of an Exojax-like syntax was added in the exomol.py file. This example introduces a new class called mdbExomol, which holds Jax arrays as attributes. This class is based on DataFrame loading and is designed to be a drop-in replacement for Exojax computations.
mport pathlib
import numpy as np
from radis.api.exomolapi import (
MdbExomol,
get_exomol_database_list,
get_exomol_full_isotope_name,
)
from radis.db.classes import get_molecule_identifier
def fetch_exomol(
molecule,
database=None,
local_databases=None,
databank_name="EXOMOL-{molecule}",
isotope="1",
load_wavenum_min=None,
load_wavenum_max=None,
columns=None,
cache=True,
verbose=True,
clean_cache_files=True,
return_local_path=False,
return_partition_function=False,
engine="default",
output="pandas",
skip_optional_data=True,
):
Here is a description of each function parameter:
molecule: The name of the molecule for which we want to retrieve data.
database: The name of the specific ExoMol database to use. If the value is None or “default”, the recommended database will be used.
local_databases: The path to the local directory where the ExoMol database files will be stored.
databank_name: The name to give to the ExoMol database.
isotope: The number of the specific isotope of the molecule for which you want to retrieve data.
load_wavenum_min and load_wavenum_max: The minimum and maximum wavenumbers to load.
columns: A list of specific columns to load from ExoMol files.
cache: A boolean indicating whether files should be cached for later use.
verbose: A boolean indicating whether informational messages should be displayed during the process.
clean_cache_files: A boolean indicating whether cached files should be removed after use.
return_local_path and return_partition_function: Booleans indicate whether local path to files and partition function should be returned in addition to data.
The function starts by checking the settings and determining which ExoMol database to use. Next, it creates an MdbExomol object to manage the ExoMol database, downloading files as needed. The local files are then loaded into a DataFrame, with specific columns if needed. RADIS-specific manipulations are performed on the DataFrame, such as renaming columns and adding additional data. Eventually the DataFrame is returned, possibly with other information if selected.
Then Erwan Pannier added an Example of an Exojax-like syntax, creating a ``mdbExoMol` class holding jax arrays as Attributes (based on DataFrame loading), so it can be used as a drop-in replacement
The example also shows how to : Compute an Exomol CH4 spectrum with Radis Load Exomol CH4 Jax arrays ready for an Exojax computation, by downloading the files to the .database local folder Load Exomol CH4 Jax arrays ready for an Exojax computation, by loading the files from the Radis’s database of (1) For 2. and 3., a local_databases= parameter allows to easily switch in between any local folder (to save on download & caching times if comparing the two codes)
Here is the code ( available on radis/exomol.py at e317c994451d3afa02b1fa63d90ae0eb546f36ed · radis/radis (github.com))
#%% RADIS-like Example
# uses fetch_exomol() internally
from radis import calc_spectrum
s = calc_spectrum(
wavelength_min=1.630e4,
wavelength_max=1.6305e4,
molecule="CH4",
isotope="1",
pressure=1.01325, # bar
Tgas=1000, # K
mole_fraction=0.1,
path_length=1, # cm
# broadening_method="fft", # @ dev: Doesn't work with 'voigt'
databank=(
"exomol",
"YT10to10",
), # Simply use 'exomol' for the recommended database
)
# s.apply_slit(1, "cm-1") # simulate an experimental slit
s.plot("xsection")
# %% Exojax like Example
class mdbExoMol:
# hardcode attribute names, to prevent typos and the declaration of unwanted parameters
__slots__ = [
"Sij0",
"logsij0",
"nu_lines",
"A",
"elower",
"eupper",
"gupper",
"jlower",
"jupper",
]
def __init__(
self,
molecule,
path,
nurange=[-np.inf, np.inf],
crit=-np.inf,
local_databases="~/exojax",
):
"""
Parameters
----------
molecule: molecule name
path : local path, mirror of ExoMol path
nurange : TYPE, optional
DESCRIPTION. The default is [-np.inf, np.inf].
crit : TYPE, optional
DESCRIPTION. The default is -np.inf.
Returns
-------
DataFrame
Examples
--------
::
mdbCH4 = mdbExoMol("CH4", '.database/CH4/12C-1H4/YT10to10/', nus, crit=1.e-30)
print(len(mdbCH4.nu_lines), "lines")
mdbCH4.elower
Available columns::
[
"Sij0",
"logsij0",
"nu_lines",
"A",
"elower",
"eupper",
"gupper",
"jlower",
"jupper",
]
"""
wavenum_min, wavenum_max = np.min(nurange), np.max(nurange)
if wavenum_min == -np.inf:
wavenum_min = None
if wavenum_max == np.inf:
wavenum_max = None
# Set-up database, download files and set-up cache files if needed
mdb = MdbExomol(
path,
molecule=molecule,
local_databases=local_databases,
nurange=[wavenum_min, wavenum_max],
)
# Get cache files to load :
mgr = mdb.get_dframe_manager()
local_files = [mgr.cache_file(f) for f in mdb.trans_file]
# Load them:
jdict = mdb.load(
local_files,
columns=[k for k in self.__slots__ if k not in ["logsij0"]],
lower_bound=([("nu_lines", wavenum_min)] if wavenum_min else [])
+ ([("Sij0", mdb.crit)] if not np.isneginf(mdb.crit) else []),
upper_bound=([("nu_lines", wavenum_max)] if wavenum_max else []),
output="jax",
)
# set attributes, accessible as e.g: mdb.nu_lines
for k in jdict.keys():
setattr(self, k, jdict[k])
nus = np.linspace(1e7 / 1.630e4, 1e7 / 1.6305e4)
# Download new ExoMol repo (in ~/exomol)
mdbCH4 = mdbExoMol(
"CH4",
".database/CH4/12C-1H4/YT10to10/",
nus,
crit=1.0e-30,
local_databases=".", # use local folder
)
print(len(mdbCH4.nu_lines), "lines")
mdbCH4.elower
# Or use RADIS's folder (# by default ~/.radisdb/exomol)
import radis
mdbCH4_2 = mdbExoMol(
"CH4",
"CH4/12C-1H4/YT10to10/",
nus,
crit=1.0e-30,
local_databases=pathlib.Path(radis.config["DEFAULT_DOWNLOAD_PATH"]) / "exomol",
)
# ... ready to run Jax calculations
The example added in the exomol.py file of RADIS demonstrates how to create an “mdbExoMol” class similar to Exojax, which uses jax arrays as attributes. This class allows loading ExoMol data as jax arrays, making them compatible with Exojax.
The example illustrates how to calculate a CH4 spectrum using RADIS by utilizing the “mdbExoMol” class. It also shows how to load the jax arrays ready for use in an Exojax computation by downloading the files into a local “.database” folder. Additionally, the example demonstrates how to load the jax arrays from the RADIS database, enabling easy switching between different local databases by specifying the “local_databases” parameter.
Toward a common API ?
Preliminary work in Radis #465
This preliminary work was done to prepare for a meeting between RADIS and ExoJax to discuss the implementation of a common API for database management.
The rationale behind this work is to maintain three layers of agnosticism to simplify the user experience:
Users should not have to worry about the specific format of the input database, such as HITRAN 2012, HITRAN 2020, HITEMP, GEISA, ExoMol, Kuruz, NIST, etc. This allows for easy integration of new libraries and ensures that all codes can benefit from all available libraries.
Users should not have to be concerned about the format in which the data is stored on disk, whether it’s Vaex’s HDF5, Pandas’s HDF5, Feather, or other formats. This flexibility enables the switch to more performant libraries when they become available, such as PyArrow.
Users should have the freedom to choose the output format used in calculations, whether it’s Pandas’s DataFrame, Jax arrays, Vaex DataFrame, or others. This allows for data retrieval in the desired format for each code, such as Jax for ExoJax, Pandas for the current RADIS implementation, or Vaex for future fully-out-of-core RADIS for extreme databases.
Radis state in May 2022
ExoJax has MdbExomol or MdbHIt . They are similar but don’t inherit from a common base ;
RADIS 0.12 has a DatabaseManager to handle HITEMP 2020 / HITRAN 2020; the ExoMol support is done with a MdBMol taken from Exojax with no link with the DatabaseManager.
RADIS implemented a DataFileManager class to handle (Vaex’s HDF5; Pandas’s HDF5, Feather) ; DatabaseManager calls DataFileManager internally
Radis Exomol api update #464 added 3 output : ’jax/vaex/pandas’ to RADIS’s DatabaseManager
Here Radis then suggest a code a demonstration :
# Test ExoMol
import radis
radis.config["MEMORY_MAPPING_ENGINE"] = "vaex" # 👉👉👉 choose "vaex", "pytables", "feather"
from radis.io.exomol import fetch_exomol
df = fetch_exomol("SiO", database="EBJT", isotope="1", load_wavenum_max=5000,
output="pandas" # 👉👉👉 choose : "pandas", "vaex", "jax"
)
print(df)
This code allows you to fetch ExoMol data for the SiO molecule from the specified database, control the output format, and print the resulting DataFrame.
The MdbExomol class is a molecular database class specifically designed for ExoMol, which is a database of molecular line lists for exoplanet and other hot atmospheres. It is a subclass of the DatabaseManager class.
The MdbExomol class has the following parameters:
path: The path for the ExoMol data directory or tag.
nurange: A wavenumber range list (in cm-1) or a wavenumber array.
margin: A margin for the wavenumber range (in cm-1).
crit: The line strength lower limit for extraction.
bkgdatm: The background atmosphere for broadening, e.g., H2, He.
broadf: If False, the default broadening parameters in the .def file are used.
Additionally, there are other parameters such as engine and skip_optional_data that control the memory mapping engine and the loading of optional data from the ExoMol definition file, respectively.
The MdbExomol class provides methods to load and manage the ExoMol data files. It converts the trans/states files to feather or HDF5 format for efficient loading. The loaded data is stored in a DataFrame with columns such as nu_lines, Sij0, A, elower, gpp, jlower, and more. These columns contain information about line centers, line strengths, Einstein A coefficients, lower state energy, statistical weight, J_lower, and other relevant parameters.
The class also provides examples of how to initialize the database, download the necessary files, and load the data. It references the original publications on ExoMol for more information.
Discussion between Radis and Exojax
The question of hosting the common API was addressed for the first time in discussion #257. It is suggested to create a new directory or module within Radis for the common API. However, it is noted that this may make development difficult if specific Radis features need to be integrated into the common API. A suggestion is made to create a separate module, such as “radis.api”, which would house the common API.
A proposed structure was discussed, where the DatabaseManager class is considered the core common API. Subclasses, such as HitranManager and ExomolManager, inherited from DatabaseManager to provide the specific functionalities for the Hitran and Exomol databases, respectively.
A suggestion was made to create a MdbHitran class (inheriting from HitranManager) and a MdbExomol class (inheriting from ExomolManager) for the ExoJAX API. These classes would have their own ExoJAX-specific methods, allowing ExoJAX to integrate with the common API.
Radis and Exojax agreed that separating the functionalities into separate folders is a good approach to avoid conflicts between Radis-specific and ExoJAX-specific features. Additionally, they suggested to add tests to ensure that ExoJAX requirements were tested within Radis without causing any issues.
It starts from the “radis.api” folder for the common API, which would be included within Radis. This approach would allow for the development of the common API while maintaining a close connection with Radis. Furthermore, the possibility of creating a new directory or module for an independent API in the future is considered, to allow the common API to become independent if needed.
In summary, the conversation revolves around the hosting and structure of the common API between Radis and ExoJAX. It is decided to create a separate module, “radis.api”, for the common API while keeping the possibility of independent development in the future. Specific classes are proposed for ExoJAX, enabling the integration of ExoJAX into the common API. Measures are taken to separate Radis-specific and ExoJAX-specific functionalities and to test ExoJAX requirements within Radis.
After this discussion, a series of measures was implemented to Radis in PR # 480
The common functions shared between Exojax and Radis were moved to the “radis/api” directory, allowing for better organization and separation of concerns.
Parts of Hitran, Hitemp, CDSD, and GEISA that were common to both Exojax and Radis were also moved to the “radis/api” directory.
The Exomol files were combined and moved to the “radis/api” directory, and tests were fixed accordingly.
The io.tools module was moved to the “radis/api/tools” directory for better organization.
Links in the documentation and docstrings were renamed to reflect the changes in the code structure.
Some missing files were added, isort errors on CI were fixed, and unclosed HDF5 files were addressed.
The Exomol documentation was updated, and error messages were improved.
The set_broadenings function was made compatible with both vaex and pandas DataFrames.
Modifications were made to the set_broadening function, including assuming a dictionary as input for unknown DataFrames and redefining the function to store values in a DataFrame.
The MdbExomol instance now includes self.molmass.
The codebase was merged with the upstream “add/common-api” branch.
Some abspath problems were fixed, and error messages were improved.
In November 2022, the version 0.14 was released and the common API with Exojax was added to Radis.
Review of the exomolapi.py code
This code is an API for accessing and using data from the ExoMol project. It includes a main class MdbExomol that allows downloading and accessing ExoMol files, as well as performing spectral calculations. The MdbExomol class provides methods for downloading files, creating PartFuncExoMol objects, and loading specific line data.
The download function allows downloading ExoMol files from a specified URL and saving them locally. The to_partition_function_tabulator function generates a PartFuncExoMol object from the ExoMol-specific partition function data.
The if __name__ == “__main__” section contains examples of using the MdbExomol class and other features of the API, including spectrum calculations using the RADIS library, and downloading and accessing ExoMol line data using the fetch_exomol function.
Thus, the ExoMol API provides tools for downloading, manipulating, and utilizing spectroscopic data from the ExoMol project, including transition files, partition functions, and calculated spectra.
Conclusion : How should I proceed to add to Radis a new database as Kurucz ?
Regarding implementing Kurucz, the structure of the exomolapi.py code can serve as a model for building a similar API for Kurucz data. I may create a class similar to MdbExomol that handles downloading Kurucz files, accessing the relevant data (such as atomic and molecular line data, partition functions, etc.), and provides methods for spectral calculations and other functionalities specific to Kurucz. I will have to adapt and modify the existing code to suit the data format and organization of Kurucz data.
In order to do so, I will review the structure of the Kurucz database and make a quick report in a next post before starting the implementation.